Tutorial 08: Floating point computations and least squares (solutions)
Finn Lindgren
Source:vignettes/Tutorial08Solutions.Rmd
Tutorial08Solutions.RmdIntroduction
In this lab session you will explore
- Speed of matrix computations
- Floating point accuracy in least squares estimation of statistical linear models
- Open your github repository clone project from Lab 2 or 4 and
upgrade the
StatCompLabpackage - During this lab, you can work in a
.Rfile, but working with code chunks in a.Rmdis highly recommended.
For measuring relative running times of different methods, we’ll use
the bench package. You won’t need to load the package with
library. Instead, check if bench::mark() runs
without errors. If it doesn’t, and it’s because the package is not
installed, then install it with install.packages("bench")
in the R Console.
Suggested code for the setup code chunk:
Numerical speed
Define the following function for computing a vector norm:
vec_norm <- function(x) {
sum(x^2)^0.5
}Try it on a simple vector where you can verify that the result is
correct, e.g.
## [1] 3.741657 3.741657We will now take a look at the computational speed of matrix operations.
First construct a vector and matrix :
Let’s check the speed difference between
and
(convince yourself that the two expressions would be equivalent in exact
arithmetic, and possibly also in floating point arithmetic). The
function t() construct the transpose of a matrix. Note that
when b is a vector (as opposed to a single-column matrix)
it is interpreted as either a column vector or a row vector, depending
on the operation. When b is a vector,
as.matrix(b) is always a single-column matrix, and
t(b) is a single-row matrix, but b %*% A is
interpreted as t(b) %*% A.
Which method for computing
is faster? Use bench::mark() to compare the different ways
of evaluating the expression. By extracting the variables
expression, median, and itr/sec
from the bench::mark() output using the
select() function, we can summarise the results in a
table.
Solution:
bm <- bench::mark(
t(A) %*% b,
t(t(b) %*% A),
t(b %*% A)
)
knitr::kable(bm %>% select(expression, median, `itr/sec`))| expression | median | itr/sec |
|---|---|---|
| t(A) %*% b | 44ms | 22.49966 |
| t(t(b) %*% A) | 10.36ms | 96.49608 |
| t(b %*% A) | 9.75ms | 100.85960 |
To save typing, define a function bench_table that does
the kable printing:
Solution:
The t(A) %*% b method is the slowest implementation.
Transposing a matrix requires copying the values to new locations.
Transposing a vector doesn’t require any copying.
For , the order in which matrix&vector multiplication is done is important. Define these new variables:
m <- 1000
n <- 1000
p <- 1000
A <- matrix(rnorm(m * n), m, n)
B <- matrix(rnorm(n * p), n, p)
b <- rnorm(p)Compare the running times of A %*% B %*% b,
(A %*% B) %*% b, and A %*% (B %*% b)).
Solution:
| expression | median | itr/sec |
|---|---|---|
| A %% B %% b | 26.76ms | 36.0692 |
| (A %% B) %% b | 26.75ms | 36.2397 |
| A %% (B %% b) | 1.82ms | 526.9319 |
The three versions should produce very similar numerical results
(bench::mark will complain if they don’t) Which method is
fastest? Why? Are the computed results the same?
Solution:
One matrix-matrix multiplication is very expensive compared with
matrix-vector multiplication. The results are numerically very close,
and one can likely express bounds for them in terms of
.Machine$double.eps and
.
For a square matrix
of size
,
compare the cost of solve(A) %*% b (Get the matrix inverse
,
then multiply with
)
against the cost of solve(A, b) (Solve the linear system
).
Solution:
bm <- bench::mark(
solve(A) %*% b,
solve(A, b),
check = FALSE # The results will differ by approximately 8e-13
)
bench_table(bm)| expression | median | itr/sec |
|---|---|---|
| solve(A) %*% b | 47ms | 20.88432 |
| solve(A, b) | 22.5ms | 42.78060 |
Numerical Least Squares estimation
You’ll now investigate some numerical issues when doing numerical least squares estimation.
Run the following code that generates synthetic observations from a linear model where , independent, .
## Set the "seed" for the random number sequences, so we can
## reproduce exactly the same random numbers every time we run the code
set.seed(1)
## Simulate the data
# x: 100 random numbers between 100 and 101
data <- data.frame(x = 100 + sort(runif(100)))
# y: random variation around a quadratic function:
data <- data %>%
mutate(y = (x - 100.5) / 5 + (x - 100.5)^2 + rnorm(n(), sd = 0.1))Let’s plot the data we have stored in the
data.frame:
ggplot(data) + geom_point(aes(x, y))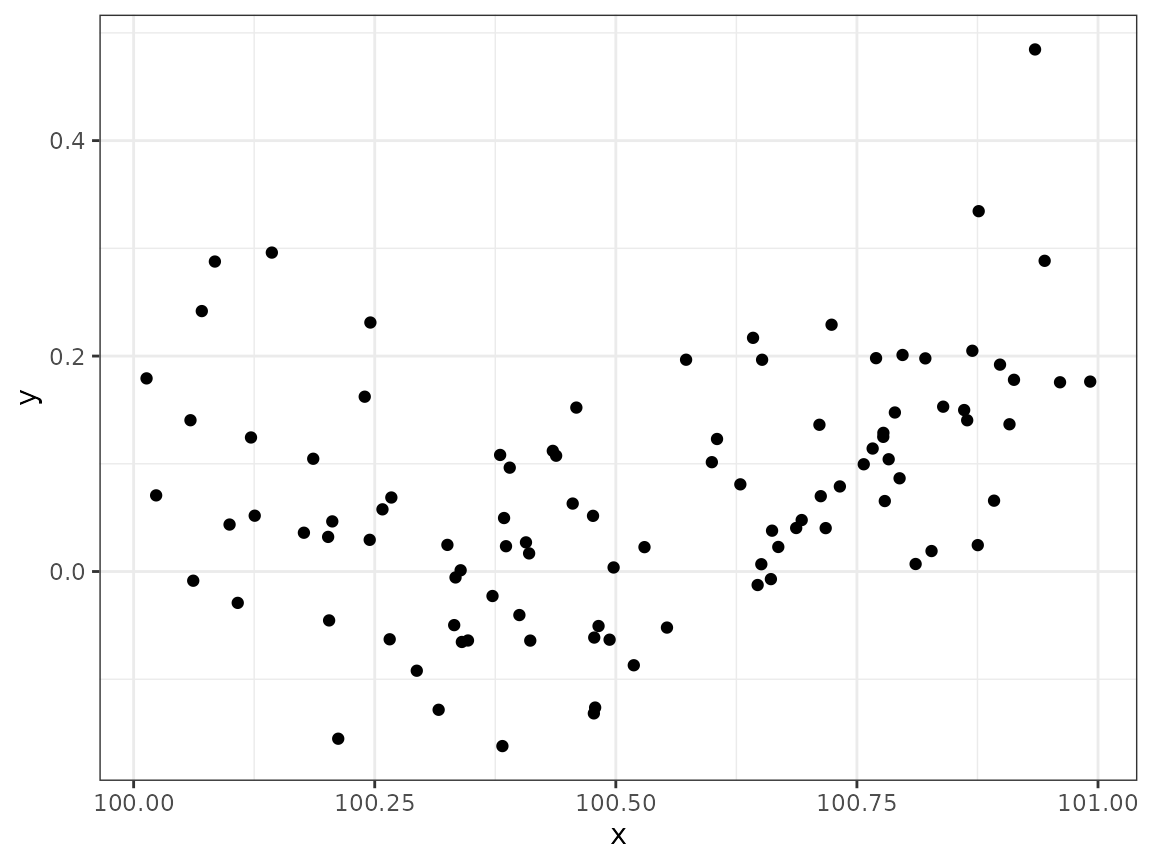
What are the true values of , , and in the standardised model formulation, Hint: Identify the values by matching the terms when expanding the initial model definition above.
Solution:
Create a beta_true vector containing
:
Solution:
beta_true <- c(10080.15, -200.8, 1)
# beta_0 = -100.5/5 + 100.5^2 = 10080.15
# beta_1 = 1/5 - 201 = -200.8
# beta_2 = 1Use these
-values
to plot the quadratic true model predictor function as a function of
,
together with the observations, and the estimate provided by
lm(). Since the observed
values are relatively dense, we don’t necessarily need a special plot
sequence of
values, but can reuse the observed values, and the model predictions for
those value can be obtained with the fitted() method. In
the lm() formula specification, use + I(x^2)
for the quadratic term. This tells lm() to first compute
x^2, and then use the result as a new covariate. The
I() method can be interpreted as “use the computed value of
this instead of other possible special meanings of the expression”.
Solution:
data <- data %>%
mutate(mu_true = beta_true[1] + beta_true[2] * x + beta_true[3] * x^2)
pl <- ggplot(data) +
geom_point(aes(x, y)) +
geom_line(aes(x, mu_true, col ="True"))
## lm manages to estimate the regression:
mod1 <- lm(y ~ x + I(x^2), data = data)
## Add the fitted curve:
pl +
geom_line(aes(x, fitted(mod1), col = "Estimated"))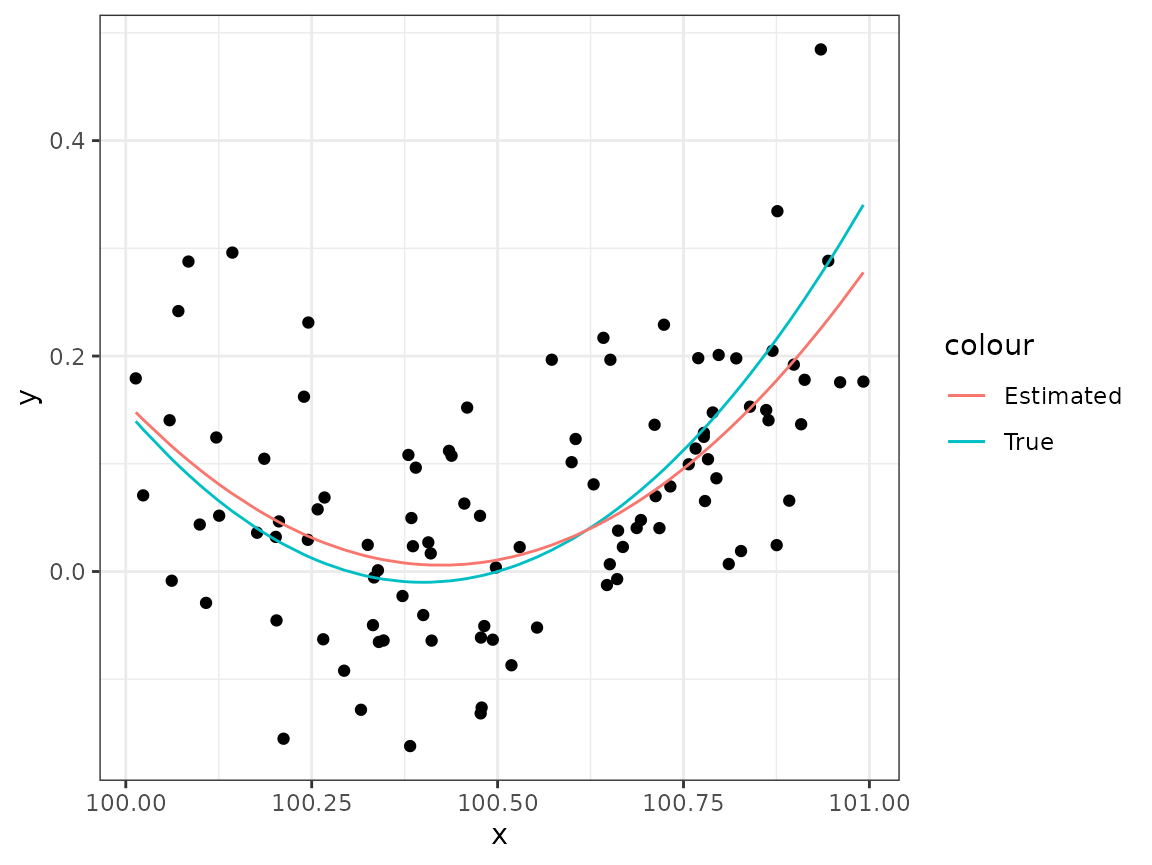
Use the model.matrix(mod1) function to extract the
matrix for the vector formulation of the model,
,
and store in X1. (See ?model.matrix for more
information). Then use the direct normal equations solve shown
in Lecture 8,
,
to estimate the
parameters. Does it work?
Solution:
X1 <- model.matrix(mod1)
# To allow the tutorial document to keep running even in case of errors, wrap in try()
try(
beta.hat <- solve(t(X1) %*% X1, t(X1) %*% data$y)
)## Error in solve.default(t(X1) %*% X1, t(X1) %*% data$y) :
## system is computationally singular: reciprocal condition number = 3.98647e-19Shifted covariate
What if the whole model was shifted to the right, so that we were given instead of the original -values? The model expression would still be able to represent the same quadratic expression but with different values. Create a new data set:
Estimate the model using lm() for this new data set.
Does it work?
Solution:
mod2 <- lm(y ~ x + I(x^2), data = data2)
ggplot(data2) +
geom_point(aes(x, y)) +
geom_line(aes(x, fitted(mod1), col = "mod1")) +
geom_line(aes(x, fitted(mod2), col = "mod2"))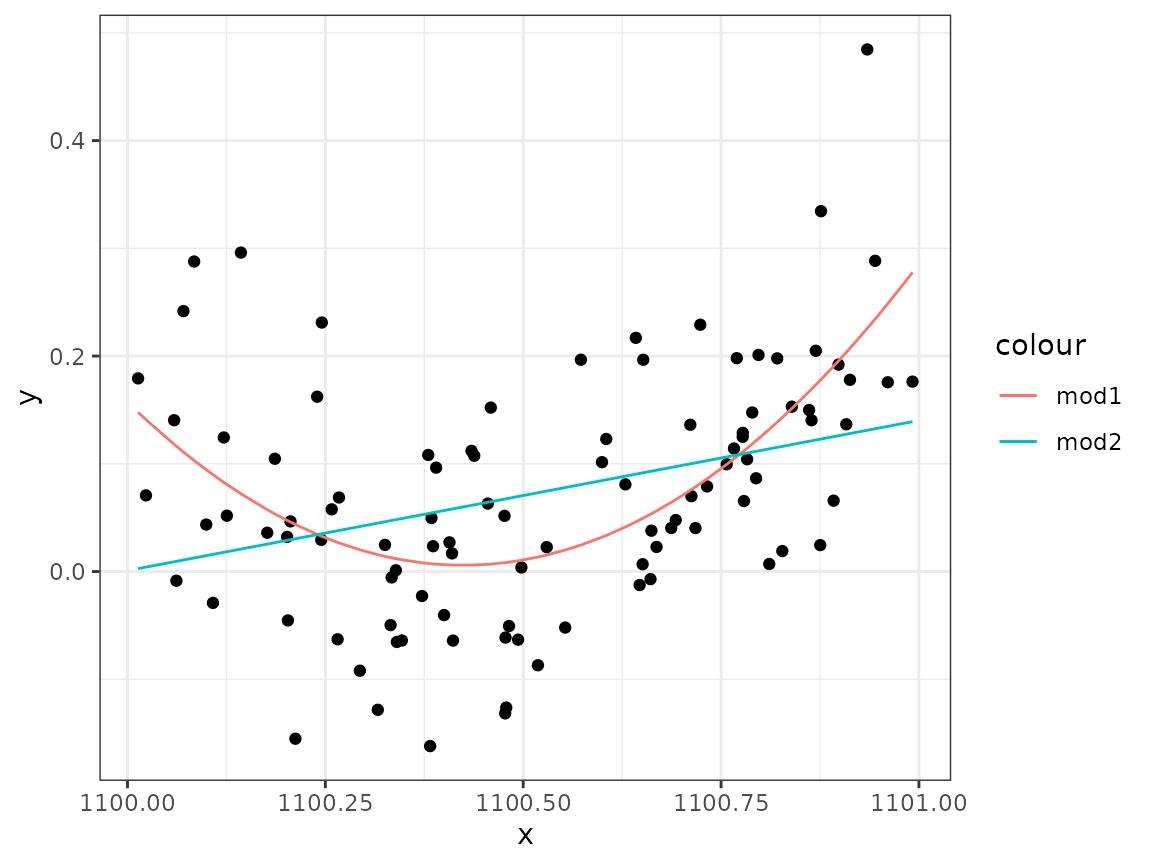
# lm() fails to fit the correct curve, without warning!lm() fails to fit the correct curve, without
warning!
The default for lm() is to allow singular models, and
“fix” the problem by removing co-linear columns of the model matrix. We
can ask it to disallow singular models with the singular.ok
argument.
Solution:
## Error in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...) :
## singular fit encountered
# Now lm refuses to compute an estimate.Define a function cond_nr() taking a matrix
X as input and returning the condition number, computed
with the help of svd (see Lecture 8 for a code example for
the condition number).
Solution:
With the help of model.matrix, examine the condition
numbers of the model matrices in the two cases from the previous tasks.
lm() computes the fit using the QR decomposition approach,
not by direct solution of the normal equations. Why was the second
lm() fit so bad?
Solution:
cond_nr(X1)## [1] 1560769713
# large, but somewhat manageable condition number
X2 <- model.matrix(mod2)
cond_nr(X2)## [1] 2.242481e+13
# very large condition number,
# but not large enough to trigger a warning!Plot the second and third columns of the model matrix X2
against each other, and use cor to examine their
correlation (see ?cor). How does this explain the large
condition number?
cor(X2[, 2:3])## x I(x^2)
## x 1 1
## I(x^2) 1 1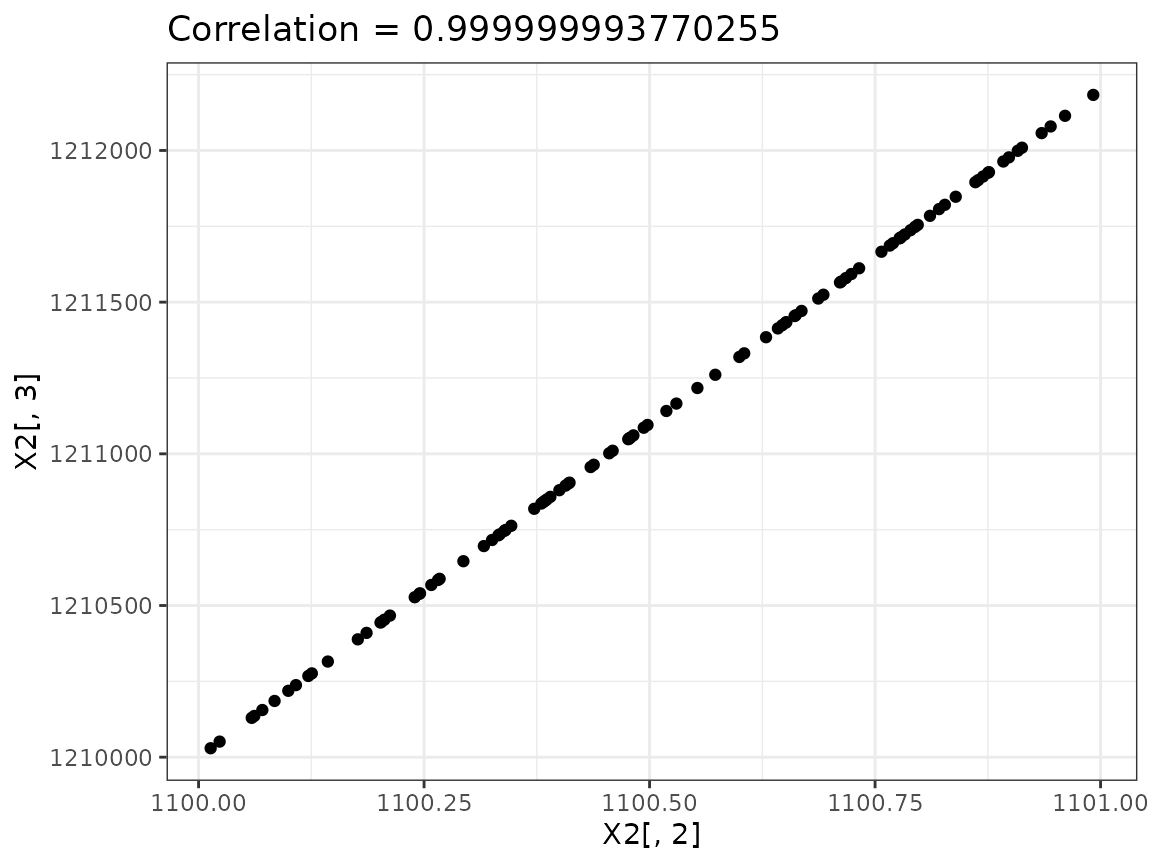
## Very close to co-linearSolution:
In both cases the columns are virtually co-linear, which leads to a very small value, and therefore a large condition number.
Since the linear model says simply that the expected value vector lies in the space spanned by the columns of , one possibility is to attempt to arrive at a better conditioned by linear rescaling and/or recombination of its columns. This is always equivalent to a linear re-parameterization.
Try this on the model matrix of the model that causes
lm() to fail. In particular, for each column (except the
intercept column) subtract the column mean (e.g., try the
sweep() and colMeans() functions, see example
code below). Then divide each column (except the intercept column) by
its standard deviation. Find the condition number of the new model
matrix. Fit the model with this model matrix using something like
mod3 <- lm(y ~ x1 + x2 + x3 - 1, data = data3)
Produce a plot that confirms that the resulting fit is sensible now.
Solution:
## Solution code:
mean_vec <- colMeans(X2[, 2:3])
sd_vec <- c(sd(X2[, 2]), sd(X2[, 3]))
X3 <- cbind(
X2[, 1],
sweep(X2[, 2:3], 2, mean_vec, FUN = "-")
)
X3[, 2:3] <-
sweep(X3[, 2:3], 2, sd_vec, FUN = "/")
data3 <- data2 %>%
mutate(x1 = X3[, 1], x2 = X3[, 2], x3 = X3[, 3])
mod3 <- lm(y ~ x1 + x2 + x3 - 1, data = data3)
ggplot(data3) +
geom_point(aes(x, y)) +
geom_line(aes(x, fitted(mod3)))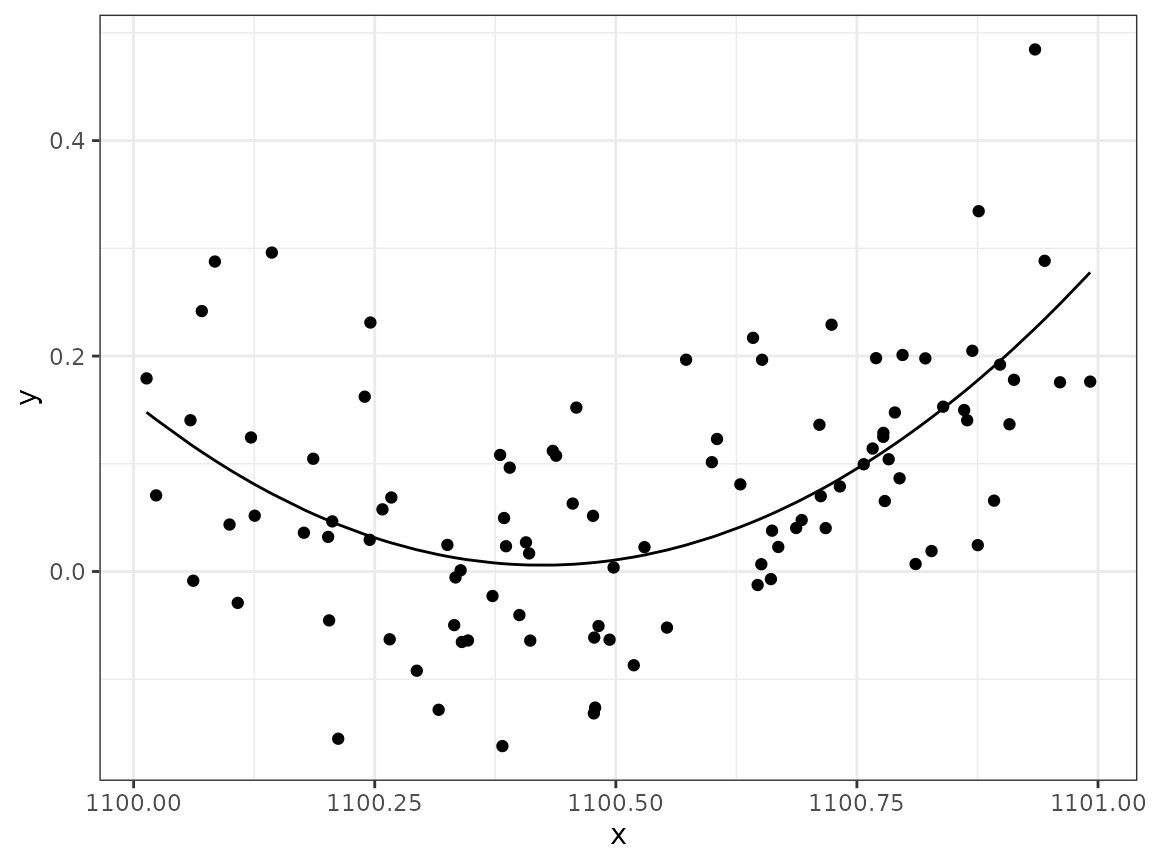
Note: If the data is split into estimation and test sets, the same
transformation must be applied to the test data, using the scaling
parameters derived from the observation data. Therefore the
scale() function isn’t very useful for this. Instead,
explicitly computing and storing the transformation information is
necessary.
Compute the condition numbers for X2 and
X3. Also compute the correlation between column 2 and 3 of
X3 (subtract
to see how close to
it is).
Solution:
c(cond_nr(X2), cond_nr(X3))## [1] 2.242481e+13 1.791760e+04## [1] -6.229745e-09 -6.229745e-09Did subtracting and rescaling help?
Solution:
Yes, the condition number was significantly reduced. The correlation between the columns is however still very close to 1, meaning that the matrix is still close to singular.
An alternative fix is to subtract the mean x value from
the original x vector before defining the quadratic model.
Try this and see what happens to the condition number and column
correlations of the model matrix. First, construct the modified
data:
Solution:
mod4 <- lm(y ~ x_shifted + I(x_shifted^2), data = data4)
ggplot(data4) +
geom_point(aes(x, y)) +
geom_line(aes(x, fitted(mod4)))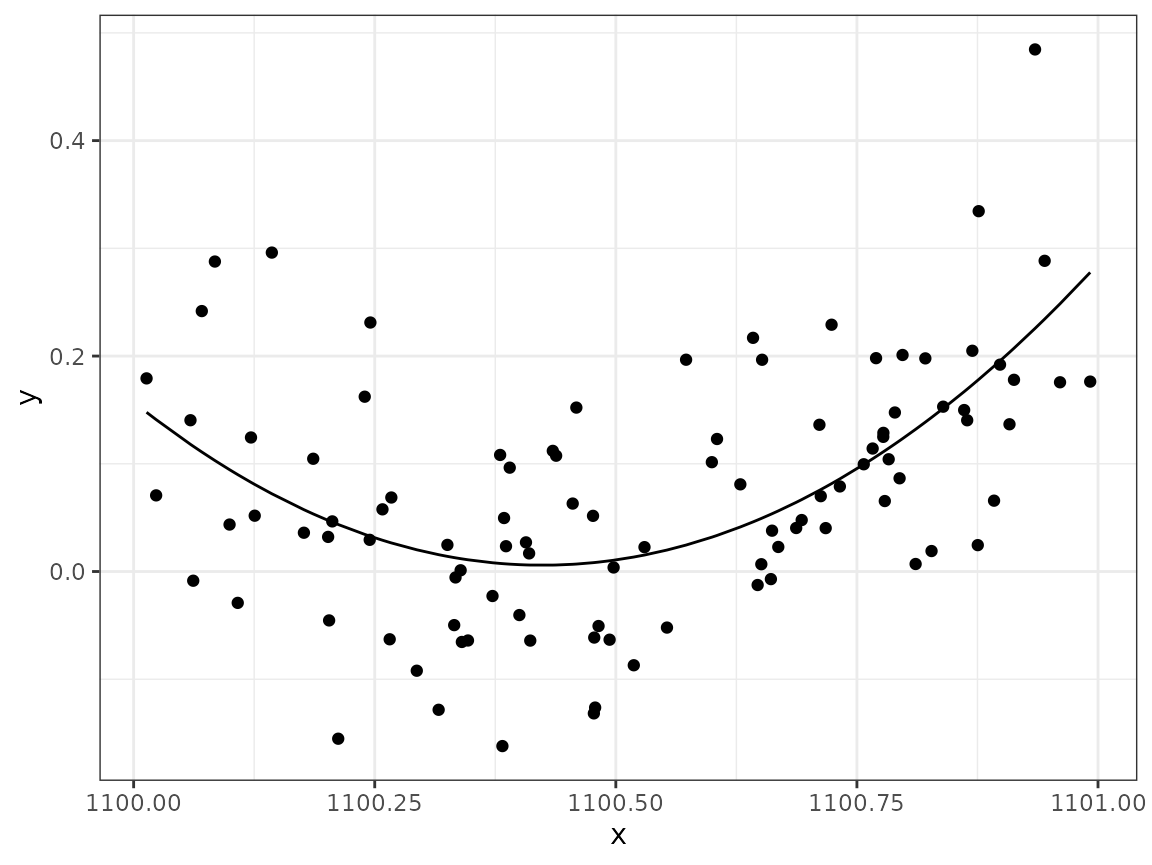
X4 <- model.matrix(mod4)
cond_nr(X4)## [1] 15.36915
cor(X4[, 2], X4[, 3])## [1] -0.09094755Different methods for modifying the inputs and model definitions require different calculations for compensating in the model parameters, and has to be figured out for each case separately. The most common effect is to change the interpretation of the parameter.